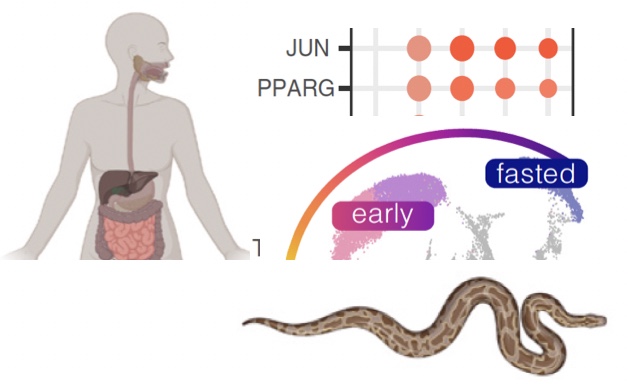
Congrats to Castoe lab PhD students Aundrea Westfall, Sid Gopalan and other coauthors for their recent paper in PNAS! This paper provides exciting new evidence for conserved intestinal regenerative growth mechanisms in vertebrates, based on single-cell RNAseq of python intestinal regeneration and comparisons with mammalian gastric bypass reprogramming.
Westfall, A.K, S.S. Gopalan, J.C. Kay, T.S. Tippets, K. Lackey, S.M. Choudhury, M.W. Pellegrino, and T.A. Castoe. 2024 Single-cell resolution of python intestinal regeneration without crypts illuminates conserved vertebrate regenerative mechanisms. Proceedings of the National Academy of Sciences USA, 121:e2405463121. PDF
Summary:
Some degree of intestinal regeneration is common across vertebrates, including humans. In mammals, the process of renewing the absorptive cells of the intestine is accomplished by stem cells that originate in intestinal crypts. Heavy-bodied snakes, such as boas and pythons, do not possess intestinal crypts, yet are able to undergo among the most extreme intestinal regeneration found in the animal kingdom. Shortly after feeding, a python’s small intestine wet mass doubles, intestinal microvillus length increases five-fold, and their oxidative metabolism increases 44-fold. Pythons therefore provide a valuable model for understanding vertebrate regenerative mechanisms that
operate independent of intestinal crypts.
We used single cell RNA sequencing to study intestinal regeneration in Pythons and found that they use conserved pathways that are also found in humans, but activate them in unique ways. Interestingly, the signaling pathways that regulate python regeneration share key similarities to those observed in humans following a specific medical intervention Roux-en-Y gastric bypass (RYGB) to facilitate weight loss and type 2 diabetes treatment. These findings highlight key fundamental links between intestinal regeneration and metabolic reprogramming, and indicate that pathways involved in python regeneration are conserved in mammals, including humans, and represent potential therapeutic targets.
Our findings also shed light on the importance of a specific intestinal cell type (BEST4+ cells) in coordinating the regeneration process. These cells are present in pythons and humans, but absent in commonly used mammalian models (e.g., mice), and act as central regulators of early phases of
regeneration by promoting lipid transport and metabolism. These findings highlight the importance and largely neglected roles BEST4+ cells likely play in human intestinal function. Together, our findings expand our understanding of vertebrate intestinal physiology and regenerative capacity with applications to metabolic and gastrointestinal disorders such as diabetes and celiac disease, as well as cancer.