Research Funding
The lab has been fortunate enough to receive grants from the NSF and NIH. Below we outline these major funded projects, and NOTE: we are now recruiting new highly motivated Ph.D. students and Postdoctoral Fellows to come and develop dissertation projects around these exciting funded projects!

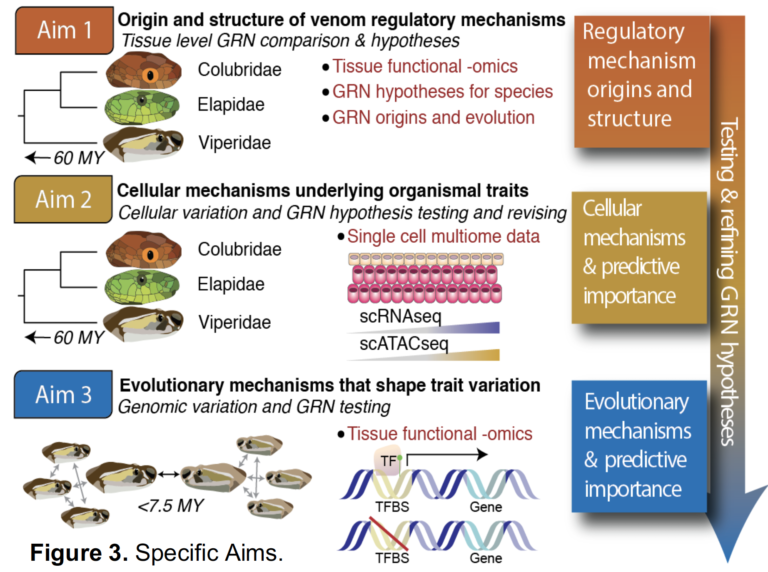
NSF-IOS: Snake venom systems as a model for inferring the structure and evolution of regulatory networks underlying organism-level physiological traits
Todd Castoe as PI, and Stephen P. Mackessy (U. Northern Colorado), Richard Adams (U. Arkansas) and Anthony Saviola (U. Colorado School of Medicine) as Co-PIs
The overarching goal of this research program is to advance the ability to understand and predict how new gene regulatory networks arise and how variation in these networks shape complex physiological traits, using snake venom as a model system. New methods developed will integrate predictive approaches and diverse functional genomic data to test mechanistic hypotheses and predict the roles of regulatory elements, trans-acting factors and other features that govern venom composition, and to test how cellular and evolutionary heterogeneity together shape organismal-level phenotypes. Key products of this work include novel approaches that leverage both single-cell variation and evolutionary variation to test hypotheses for the roles of gene regulatory components that will be broadly applicable to any eukaryotic system. By providing new insights into how snake venom variation is regulated, the project will have broad ramifications for improving global treatment of snakebite, with potential to impact millions globally.

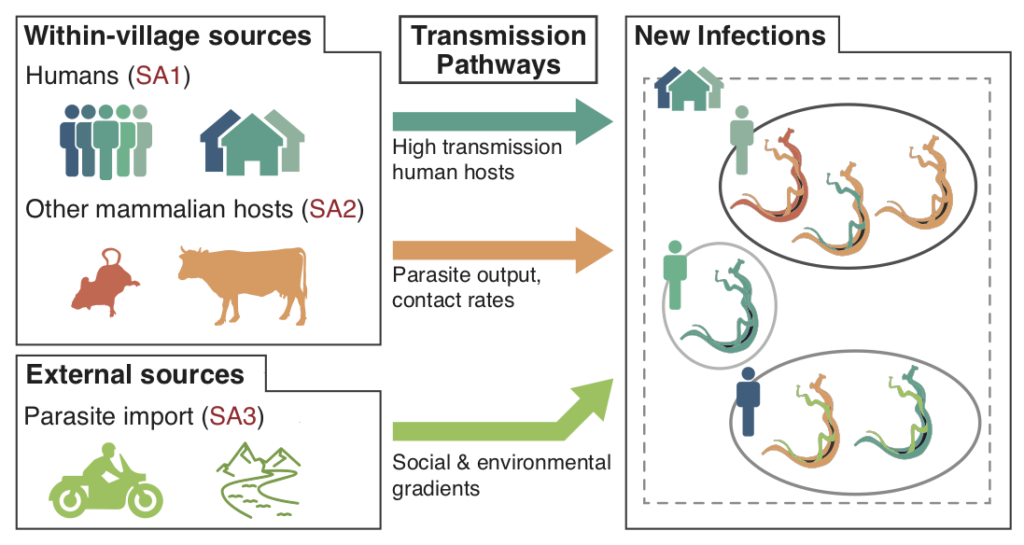
NIH-NIAID: Schistosomiasis at the edge of elimination: characterizing sources of new infections in residual transmission hotspots.
Elisabeth Carlton (PI, U Colorado), David Pollock (Co-I, U Colorado), and Todd Castoe (Co-I, UTA)
Over 200 million people are affected by schistosomiasis worldwide. The proposed research will leverage new data collection and analysis tools to understand why schistosomiasis persists in some areas where extensive control measures have been implemented. The proposal will elucidate how infections spread locally and migrate regionally, and how the parasite responds to control measures. Using cutting-edge genomic approaches, the research program will provide unprecedented insight into the detailed patterns of transmission across hosts, across geographic areas, and through time. This will help understand how to prevent infections and advance efforts to achieve permanent reductions in schistosomiasis and other human helminthiases.

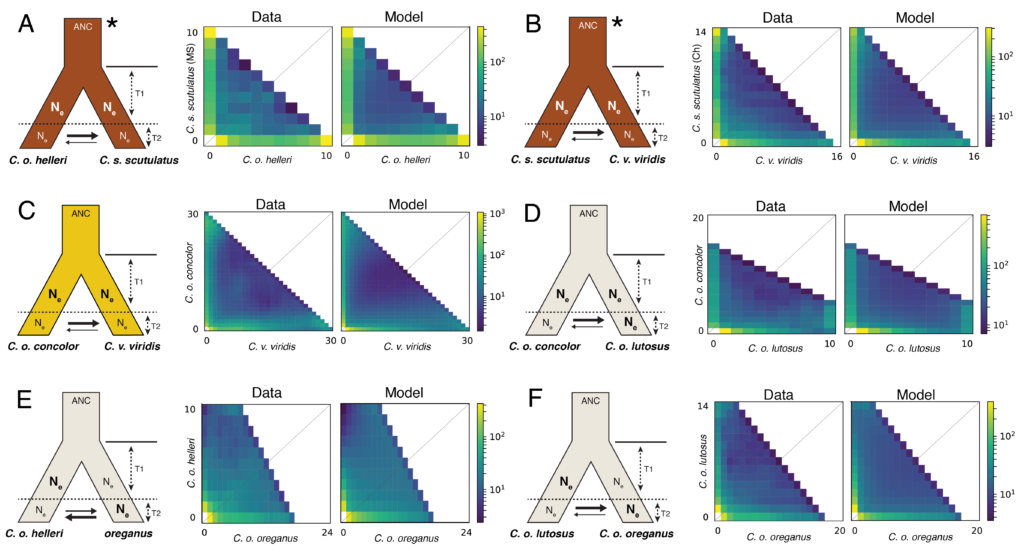
NSF-DEB: Systematics, introgression, and adaptation in Western Rattlesnakes: a model system for studying gene flow, selection, and speciation. (completed, 2023)
Todd Castoe as PI, and Stephen P. Mackessy (U. Northern Colorado), Jesse Meik (Tarleton State U.) and Matthew Fujita (UTA) as Co-PIs
The overarching goals of this research program are to understand the interaction of admixture and selection in speciation, and to leverage an empirical system to test how these processes may influence coalescent-based species delimitation methods. This research program will focus on the Western Rattlesnake species group (Crotalus viridis species complex and its sister taxon, C. scutulatus; collectively Cvos hereafter) as a study system. This recently diverged species complex has offered major challenges for systematics, yet provides an ideal model system for studying speciation and species delimitation. This integrated study will combine genomic and phenotypic data to delimit species, inform species delimitation approaches, and provide new genome-scale insight into the process of speciation. This research program will also test hypotheses about the repeatability of patterns selection in the processes of speciation and resistance to gene flow and hybridization. The specific aims of this proposal are to: (1) integrate genome-wide sampling and ecological niche modeling to infer population structure and test hypotheses of gene flow among lineages of Cvos, (2) analyze selection in contact zones between putative taxa by integrating niche modeling, genotypic, and phenotypic data to test hypotheses regarding divergence and selection in the speciation process, and (3) develop and test new approaches for delimiting species that more appropriately handle admixture and selection, and apply these methods to test taxonomic hypotheses for the Cvos complex.
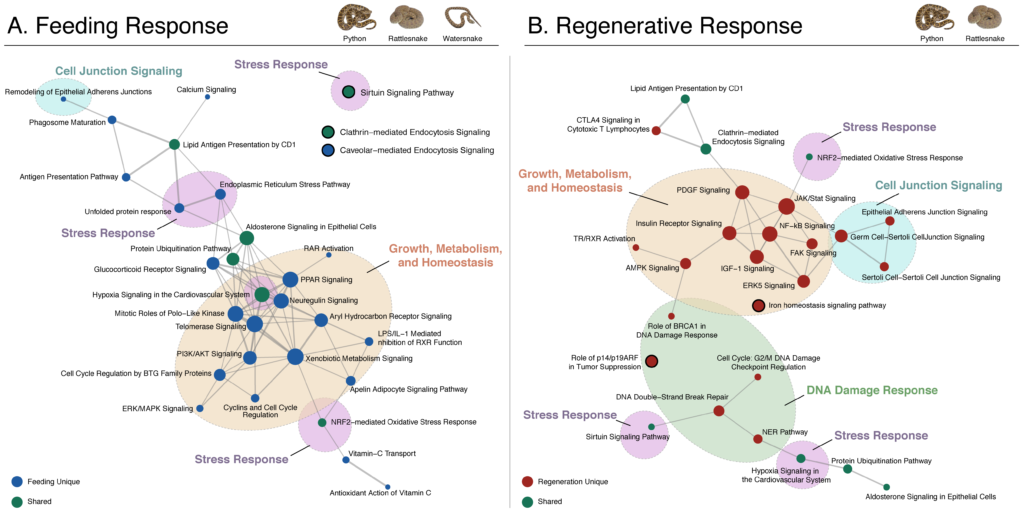
NSF-IOS: Collaborative Research: Integrated mechanisms underlying the regulation of Intestinal form and function. (completed, 2023)
Todd Castoe and Stephen Secor (U. Alabama) as PIs, Saiful Chowdhury (UTA-Chemistry and Biochemistry) as Co-PI
Vertebrates exhibit a broad range of physiological capacities to alter intestinal performance that are adaptively linked to their feeding habits. For example, species that naturally experience long episodes of fasting (e.g., sit-and-wait foraging snakes) experience rapid upregulation of intestinal form and function with feeding, and subsequent intestinal atrophy and downregulation following the completion of digestion. In contrast, frequently feeding species (e.g. active-foraging snakes) experience only modest change in intestinal form and function with feeding or fasting. Currently unknown are the proximate cellular and molecular mechanisms that underlie the structural and functional flexibility of the intestine, and whether such mechanisms are shared across vertebrates that similarly widely or narrowly regulate intestinal performance. For the widely regulating Burmese python it is known that feeding triggers the differential expression of over 1000 intestinal genes, including those involved in regulatory, cytoskeletal, trafficking, transporter, and hydrolase expression programs. By leveraging the extreme range in intestinal responses exhibited by snakes and other vertebrates and recently available genomic resources, this research program will identify the underlying mechanisms of intestinal flexibility and test if these mechanisms are shared or unique across lineages and regulatory phenotypes. These broad questions will be addressed by pursuing three aims: (1) identify the cellular and structural mechanisms that underlie the modulation of intestinal form, and whether form dictates the regulation of intestinal function; (2) link transcriptional and post-translational mechanisms to phenotypic changes in intestinal structure and function; and (3) test whether shared or unique sets of molecular mechanisms drive similar phenotypic responses among vertebrates. Ultimately, this research will identify the signaling and structural mechanisms by which vertebrates modulate intestinal form and function.