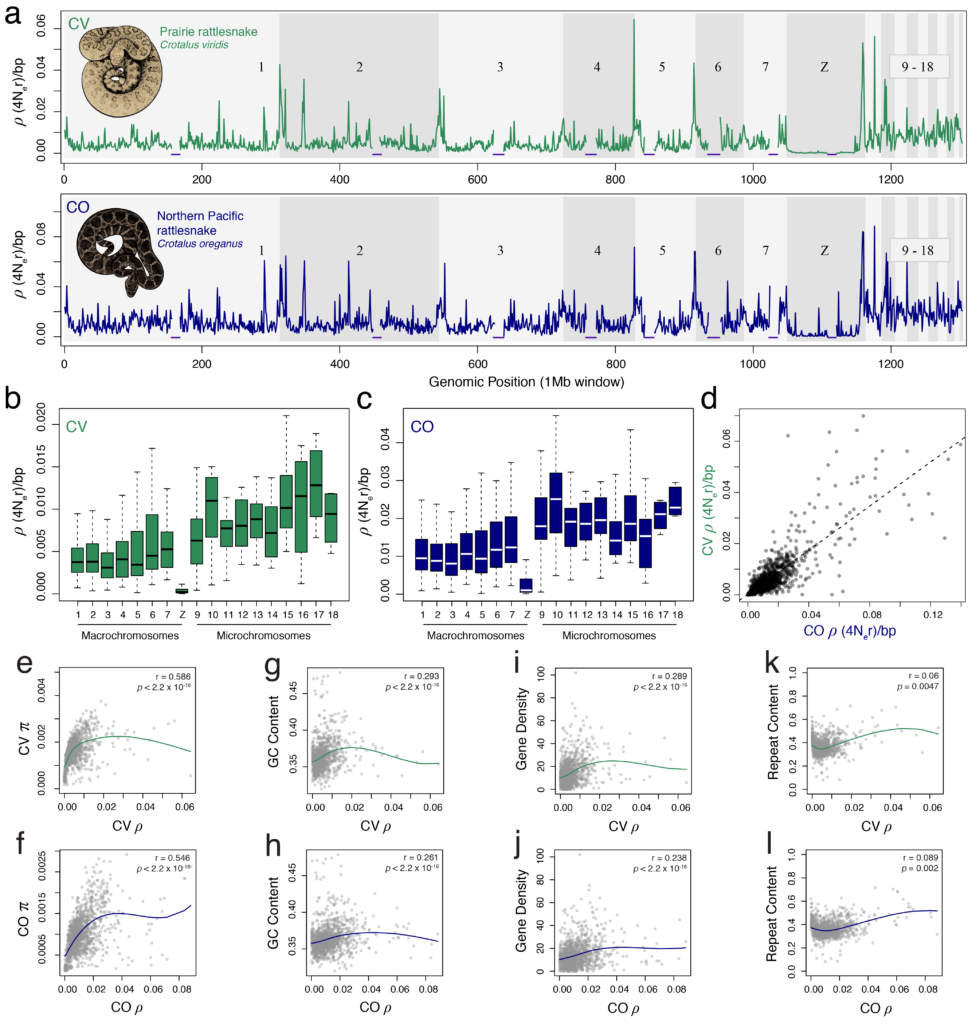
Our paper on snake recombination has been accepted at Molecular Biology and Evolution! Using whole genome data from populations of two rattlesnake species, we characterize the genomic recombination landscape and investigate the relevance of PRDM9 in snake recombination. We find evidence that snakes are unique among vertebrates in that PRDM9 is involved in meiotic recombination, yet recombination hotspots are concentrated in functional regions associated with genes and their regulators.
Our results provide the first recombination maps for snakes, and illustrate unique features of snake genome evolution and the value of further investigation into snake and squamate recombination.
Congratulations to Drew and all co-authors!
Schield, D.R., G.I.M. Pasquesi, B.W. Perry, R.H. Adams, Z.L. Nikolakis, A.K. Westfall, R.W. Orton, J.M. Meik, S.P. Mackessy, and T.A. Castoe. 2020. Snake recombination landscapes are concentrated in functional regions despite PRDM9. Molecular Biology and Evolution. PDF
The rattlesnake recombination maps are available here.
Abstract
Meiotic recombination in vertebrates is concentrated in hotspots throughout the genome. The location and stability of hotspots have been linked to the presence or absence of PRDM9, leading to two primary models for hotspot evolution derived from mammals and birds. Species with PRDM9-directed recombination have rapid turnover of hotspots concentrated in intergenic regions (i.e., mammals), while hotspots in species lacking PRDM9 are concentrated in functional regions and have greater stability over time (i.e., birds). Snakes possess PRDM9, yet virtually nothing is known about snake recombination. Here we examine the recombination landscape and test hypotheses about the roles of PRDM9 in rattlesnakes. We find substantial variation in recombination rate within and among snake chromosomes, and positive correlations between recombination rate and gene density, GC content, and genetic diversity. Like mammals, snakes appear to have a functional and active PRDM9, but rather than being directed away from genes, snake hotspots are concentrated in promoters and functional regions – a pattern previously associated only with species that lack a functional PRDM9. Snakes therefore provide a unique example of recombination landscapes in which PRDM9 is functional, yet recombination hotspots are associated with functional genic regions – a combination of features that defy existing paradigms for recombination landscapes in vertebrates. Our findings also provide evidence that high recombination rates are a shared feature of vertebrate microchromosomes. Our results challenge previous assumptions about the adaptive role of PRDM9 and highlight the diversity of recombination landscape features among vertebrate lineages.